Computer-based clinical coding activity analysis for neurosurgical terms
Article information
Abstract
Background
It is not possible to measure how much activity is required to understand and code a medical data. We introduce an assessment method in clinical coding, and applied this method to neurosurgical terms.
Methods
Coding activity consists of two stages. At first, the coders need to understand a presented medical term (informational activity). The second coding stage is about a navigating terminology browser to find a code that matches the concept (code-matching activity). Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT) was used for the coding system. A new computer application to record the trajectory of the computer mouse and record the usage time was programmed. Using this application, we measured the time that was spent. A senior neurosurgeon who has studied SNOMED CT has analyzed the accuracy of the input coding. This method was tested by five neurosurgical residents (NSRs) and five medical record administrators (MRAs), and 20 neurosurgical terms were used.
Results
The mean accuracy of the NSR group was 89.33%, and the mean accuracy of the MRA group was 80% (p=0.024). The mean duration for total coding of the NSR group was 158.47 seconds, and the mean duration for total coding of the MRA group was 271.75 seconds (p=0.003).
Conclusion
We proposed a method to analyze the clinical coding process. Through this method, it was possible to accurately calculate the time required for the coding. In neurosurgical terms, NSRs had shorter time to complete the coding and higher accuracy than MRAs.
Introduction
Recently, the ability to collect and analyze data has developed, and researches using so-called "big data" have been actively conducted in various fields [1]. In the medical area, various studies using big data also have been attempted. However, differences in clinical coding systems and data structures are major barriers to such researches [2]. Therefore, researches on data standardization using a tool such as a common data model is being carried out [3]. In our country, researches on standardizing medical data are also conducted by the government [2].
However, maintaining such standardized medical data requires human and financial resources in hospitals, and there have been challenges regarding accuracy, coding variation, quality assurance and so on [4-6]. Recently, the adoption and use of electronic health records (EHRs) system has been increasing worldwide. Thus, it has become easier to collect medical data. However, most EHRs are not interchangeable [7]. In the neurosurgical fields, the incidence of diseases is relatively low, and there are many surgical techniques that have relatively low frequency. Thus, medical data before EHRs is also very important in the neurosurgical field.
It is not possible to measure how much activity is required for individuals to understand a medical data objectively or how much time is needed to search codes when they already know the meaning of medical data. Such information will be very important in predicting the costs of future research. We will also be able to identify problems with clinical coding. The purpose of this study is to propose a clinical coding activity analysis method using the computer mouse, and to analyze the results after coding of neurosurgical terms using this method.
Material and methods
1. Collection of neurosurgical data
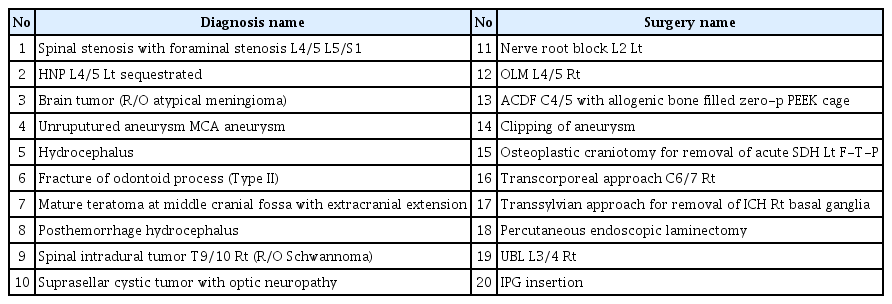
We selected 1,071 patients who was admitted to the neurosurgical department for a year at our institute. We reviewed the name of the surgery and the diagnosis. From this pool of terms, we randomly selected 10 names of the diagnosis and 10 names of the surgery using “Rand function” of Microsoft Office Excel 2013 (Microsoft Co., Redmond, WA, USA). Selected terms are listed in Table 1.
2. Systematized Nomenclature of Medicine – Clinical Terms
The Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT) is an international clinical terminology that can facilitate interoperability by capturing clinical data in a standardized manner. The International Health Terminology Standards Development Organization was established to maintain and promote SNOMED CT as a clinical reference terminology. Many countries have designated SNOMED CT as the preferred clinical reference terminology for use in EHRs [8]. However, it is not used in our country. We used SNOMED CT, which coders participating in this study have never used.
3. Clinical coding activity
In this study, clinical coding activity consists of two stages. In the first stage, the coders need to understand a presented medical term (informational activity). They use the internet search engine to understand the meaning of terms or abbreviations during information activity. The second stage is about navigating the terminology browser to find a code that matches the concept (code-matching activity). In order to evaluate the accuracy of these activities, a senior neurosurgeon who has studied SNOMED CT has analyzed the accuracy of input coding.
This coding activity was carried out by five neurosurgical residents (NSRs) as domain experts and five medical record administrators (MRAs) as coding experts. The NSRs consisted of two fourth-year residents, two third-year residents, and a second-year resident. And, the career of MRAs was at least 5 years. However, both of the groups had very limited experience with SNOMED CT and CliniClueXplore, which was used as the browser of SNOMED CT. Before the experiment, we briefly gave instruction on how to use CliniClueXplore and SNOMED CT in general to each test subject. The time required for introduction was about 20 minutes. During the test, there was no more explanation or instruction for understanding the meaning of given terms. The randomly collected diagnosis and surgery terms were presented to each participant to perform the clinical coding activity.
4. Computer-based clinical coding activity analysis
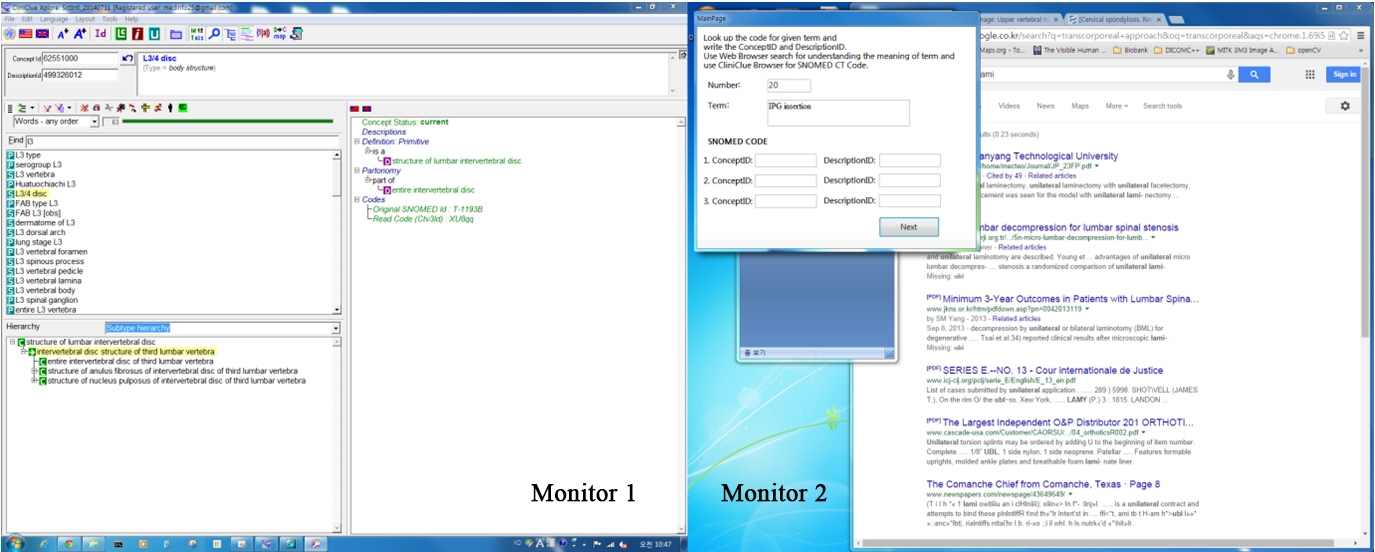
The user’s computer-based clinical coding activity analysis (CBCCAA) was implemented as follows. A computer application to record the trajectory of the computer mouse and record the usage time was programmed. And user interface was built with Microsoft Office Access 2007 (Microsoft Co.), which could store terms and codes. On the first page of the question window, simple instructions for the experiment and input fields for demographic information were placed. When the participants clicked the ‘Start’ button, first term could be seen and the participants could enter SNOMED CT codes into the textbox of the question window (Fig. 1). At the same time, our program would start measuring the time that was spent on each activity. When the participant clicked the ‘Next’ button, the next term was presented, and the time taken for each individual term was recorded.
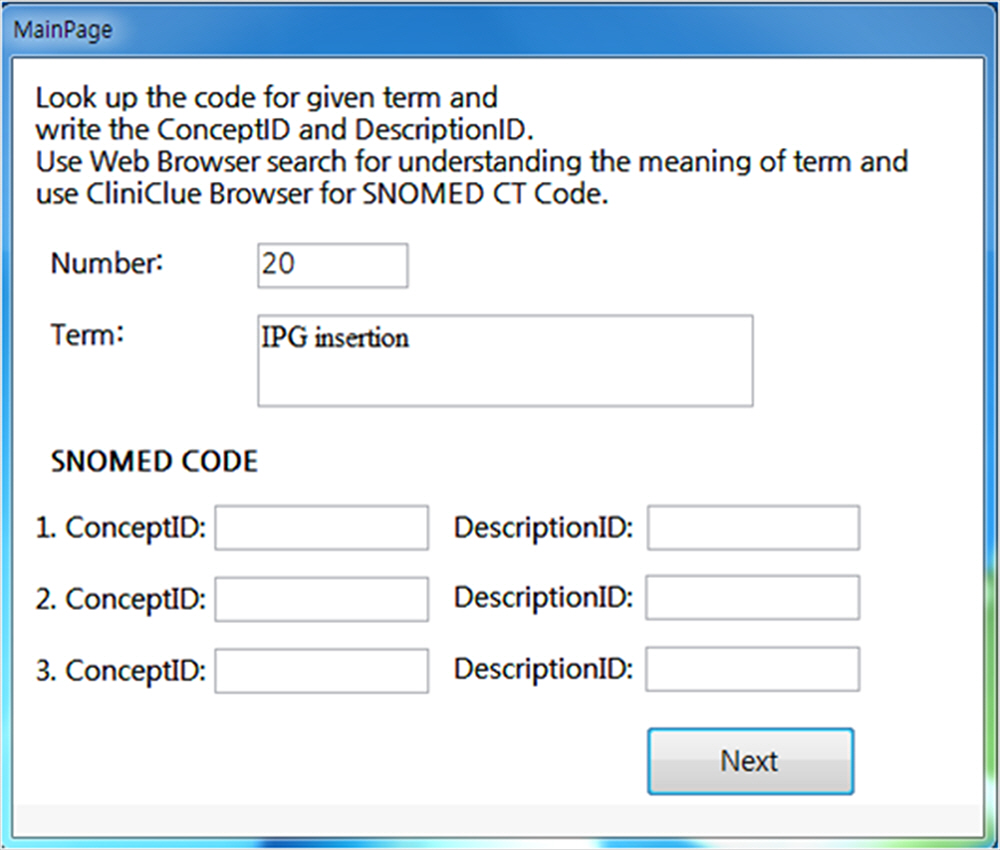
The first page of question window, which shows the term to be coded and input fields for SNOMED code. SNOMED, Systematized Nomenclature of Medicine.
With this set up, we measured the length of each user’s computer activity with our application. As we used two computer monitors to capture mouse movements, the participant could keep the SNOMED CT browser at maximum size at all times, so that we could easily analyze each user’s activity on each browser (Fig. 2). The computer activity was decided by the location of mouse clicks and wheel events. If those mouse activities were developed in the web browser, the length of the activity was assigned into the informational activity, because the user’s intention was trying to understand the meaning of a given term using web browsing. Meanwhile, the code-matching activity would be the length of clicking and wheel events on the SNOMED CT browser.
5. Statistical analysis
Statistical analysis was performed using SPSS version 18.0 (SPSS Inc., Chicago, IL, USA). Clinical coding activity was compared using an independent t-test. Probability values of less than 0.05 were considered to be statistically significant.
Results
All coding activities analyzed are listed in Table 2. In the NSR group, the most inaccurate term for coding was term 1 (spinal stenosis with foraminal stenosis L4/5 L5/S1), and the most time-consuming term for coding activity was term 16 (transcorporeal approach C6/7 right [Rt]). On the other hands, in the MRA group, the most inaccurate term for coding was tem 16 (transcorporeal approach C6/7 Rt) and term 20 (implanted pulse generator insertion), and the most time-consuming term for coding activity was term 7 (mature teratoma at middle cranial fossa with extracranial extension). The accuracy and total coding time were inversely proportional (Pearson’s correlation coefficient=–0.438, p-value=0.005).
The CBCCAA illustrated the characteristics of two coder groups. The accuracy of coding activity in the NSR group is higher than the MRA group (p=0.024). The mean duration of informational activity (p<0.001) and total duration of coding activity (p=0.003) in the NSR group was shorter than the MRA group. The mean duration of code-matching activity in the NSR group was also shorter than the MRA group, but there was no statistical significance (p=0.07) (Table 3). In the NSR group, the informational stage was significantly shorter than code-matching stage, but there was no statistical significance in the MRA group (Table 4).
Analysis of coding activity according to diagnosis versus surgery name was performed, but there was no statistically significant difference. Coding according to subspecialties of neurosurgical disorders was also analyzed. In both brain and spine part, the mean duration of informational activity and total duration of coding activity in the NSR group was shorter than the MRA group (Table 5).
Discussion
Recently, as interest in big data, deep learning, and artificial intelligence has increased, interest in data collection has increased. In order to receive compensation, the code related to the health insurance had to be continuously input, and many studies using this data have been performed [9]. However, such data are limited, and new data collection is required depending on the content of the study. For clinical data collection and standardization, coding the data at each hospital is very important. Understanding and code-searching are the two main stages of medical coding. We tried to use these two particular concepts to analyze clinical coding activity.
In this study, we provided a method to evaluate coding activity in an objective way. A benefit of this method is that the parameters are produced with numeric values that can be measured by computers. Thus, we can compare parametric results quantitatively, and can analyze the efficiency according to the coder and kinds of terminology. When we reviewed the data in this study, coding activities were very varied greatly, depending on the coders. The NRS group did more efficient coding than the MRA group. The coding time was significantly shorter in the information stage, and thus the total time was shortened (Table 3). The accuracy of coding was higher in NSR group, probably because they understood neurosurgical terms better than the MRAs did. We expected that the MRA group would take less time for code-matching, because they specialized in coding. However, there was no significant difference in the code-matching time between the NSR group and the MRA group. In the healthcare field, there are various type of occupations (physicians, dentist, nurses, MRA, clinical laboratory technologists, radiologic technologists, etc.) and costs are different. Using the method presented in this study, it can be expected which job group will be efficiently at clinical coding data.
We also analyzed coding processes by each stages. In the NSR group, the informational stage was shorter than code-matching stage. As mentioned previously, a short informational activity means that the coder has no difficulty with understanding the terms. On the other hands, long code-matching activity refers to problems matching concepts to codes in the terminology browser. Inefficient browsers can be more time-consuming in code-matching activity. Thus, if this phenomenon persists, the browser used for coding should be improved. We also analyzed the coding activity according to the kind of terminology. Neurosurgical diseases and surgery were classified into brain and spine. In both brain and spine part, coding activity of the NSR group was effective than the MRA group.
In this study, we can confirm that the coding time was longer for terms that had low coding accuracy, because less-understood terms took longer to find. For example, a term that was difficult to code in both groups was “transcorporeal approach C6/7, Rt,” because this is that this surgery has been proposed relatively recently, and is called by various names [10].
Although the coders could start the test by pressing a button, the coder’s activity was measured by the length of activity on the designated window. Because the computer mouse logging function automatically evaluates the coding process, there is no manual evaluation to categorize the coder’s activity which may lead to subjective results. Since we are heading towards a computerized coding environment, those parameters need to be evaluated by computer-based tools as well [11]. As far as we know, this is the first study to analyze the clinical coding process by means of computer activity. By this approach, we can clarify practical problems with coding activity and analyze proper solutions to such issues.
Notes
No potential conflicts of interest relevant to this article were reported.